Cells perform computations using networks of interacting molecules. It is tempting to think of these networks like electronic circuits, but in reality their components are under vastly different physical constraints. Molecules in cells are produced and destroyed at random times, are subject to large fluctuations in their numbers, and must execute random walks through crowded environments in order to interact. How can anything reliable be computed under such conditions?
The Mugler Group aims to answer this question using a wide range of theoretical and computational tools, including stochastic modeling, statistical physics, and information theory. We are tackling problems that range from the molecular to the multicellular level, often in collaboration with experimental groups, with the goal to create a better understanding of biological systems and help combat diseases such as cancer.
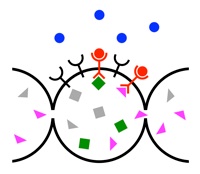 |
Collective sensing by communicating cells
Cells from bacteria to amoebae to human tissue use cell-to-cell communication to coordinate behavior. One of the most important behaviors for cells is sensing chemicals in their environment. In collaboration with experimentalists, we are finding that cells use communication to make sensing more precise. For example, clusters of cells use communication to detect shallower chemical gradients than single cells. Using tools from statistical physics, we are developing a unified theory of collective sensing.
See publications 22, 23, 27, 29, and 30.
|
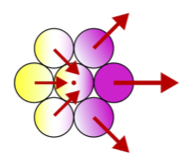 |
Collective migration and metastatic invasion
Metastasis is the deadliest stage of cancer. Metastasis begins when tumor cells invade the surrounding tissue. In many cancer types, invasion is collective, occurring via cell clusters instead of single cells. Collective invasion is one of many examples of coordinated cell migration. In collaboration with experimentalists, we are exploring collective migration using theory, simulation, and microfluidic experiments. We aim to provide a physical understanding of collective migration, as well as advance our grasp on processes that allow cancer to grow and spread.
See publications 24, 28, 33, 39, and 44.
|
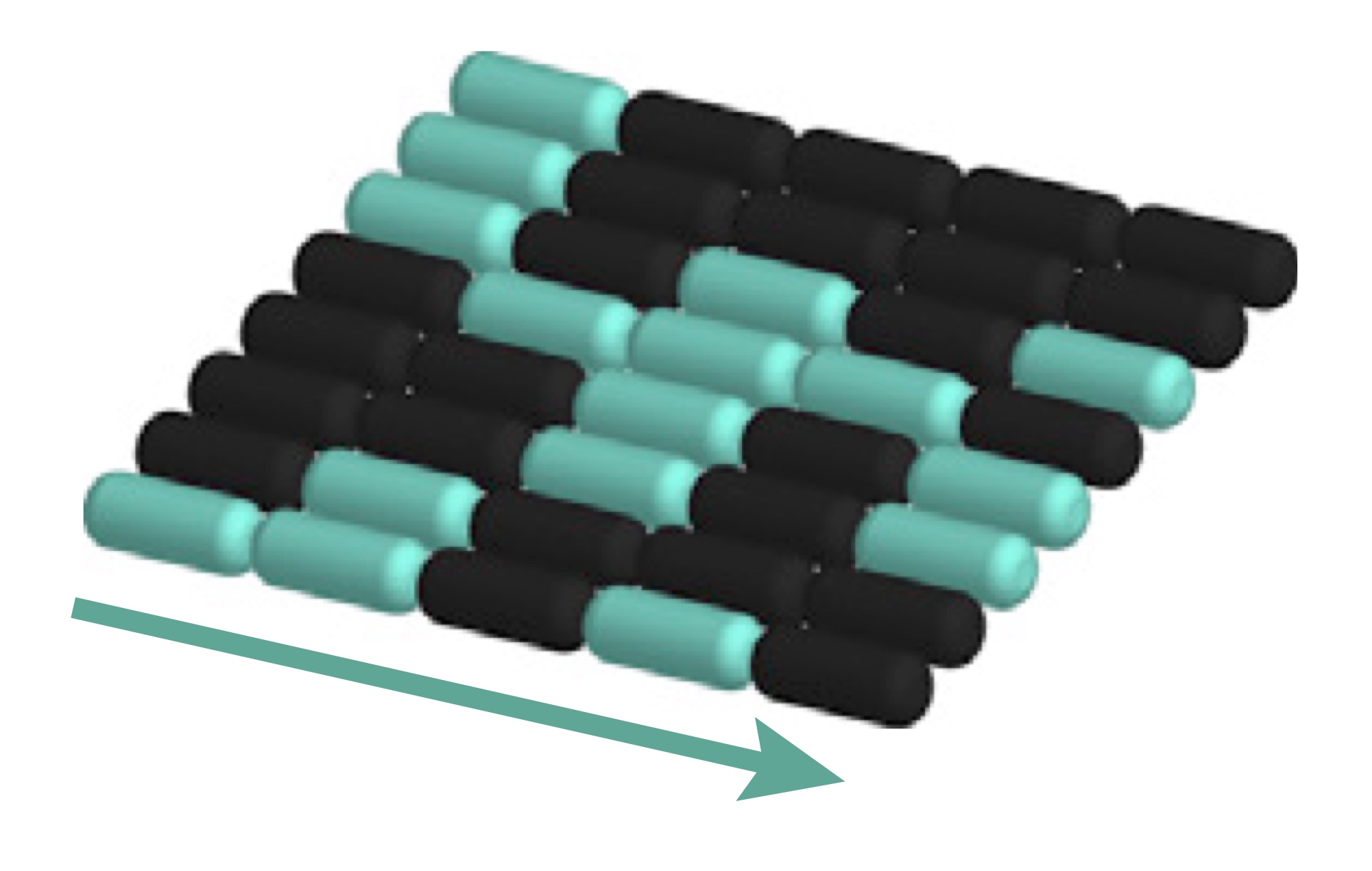 |
Signal propagation in heterogeneous communities
Cells within large communities transmit signals over long distances in order to coordinate behaviors. However, sometimes signal propagation comes at a cost. Working with experimental collaborators, we are investigating the costs and benefits of signal propagation in bacterial biofilms. We have discovered that not all bacteria participate in the signaling, and that the ones that do are arranged in a way that is well described by percolation theory, a branch of statistical physics that describes coffee filtering and crack formation.
See publications 37, 42 and 43.
|
 |
How cells tell time
Proper timing is essential for many biological functions. Even single cells can tell time. In collaboration with experimental groups, we are investigating cells that position themselves at the correct location within a developing organism by stopping their migration after a certain amount of time. We are finding that particular strategies at the genetic level can reduce noise and enhance timing precision.
See publications 35 and 45.
|
 |
Criticality in biochemical networks
Feedback in cell signaling networks leads to bifurcation points in the network dynamics. These bifurcation points share many properties with critical points in systems from statistical physics, but the implications for cell behavior are poorly understood. We are investigating the critical behavior of single- and multicellular biochemical models to understand the resulting tradeoffs for cell signaling and sensing.
See publications 38, 41, and 46.
|